LLNL’s Biological and Environmental Research (BER) program focuses on climate change resilience and energy security, contributing to both the national security mission at Lawrence Livermore and the Office of Science’s emphasis on basic science.
Using unique laboratory, field, and computational capabilities, LLNL researchers discover the underlying biology of plants and microbes as they respond to and modify their environments. On a larger scale, BER observes and models Earth system variability and change to understand and ultimately predict interactions among the planet's physical, chemical, and biological processes.
Our research portfolio includes:
- Development of exascale-capable Earth system models.
- Analysis of standardized climate and Earth system model simulations from the world's leading modeling centers.
- Investigation of cloud and aerosol physics and atmospheric chemistry.
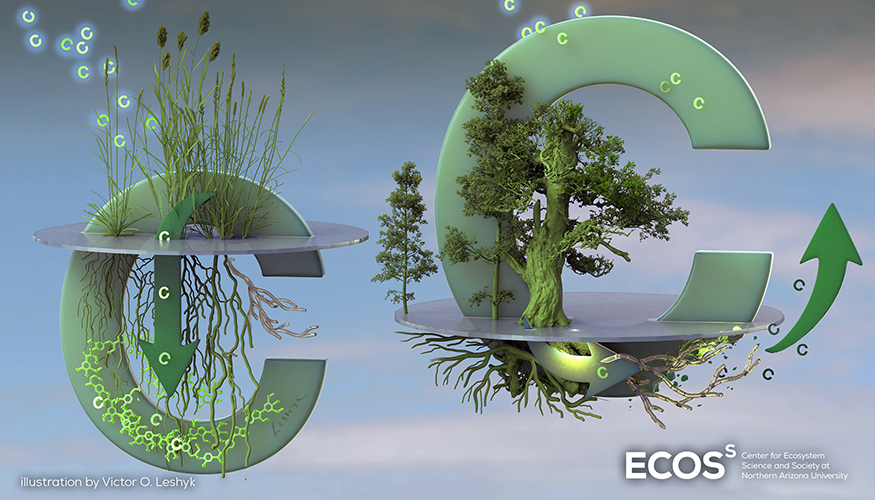
- Modeling biogeochemical processes at environmentally relevant timescales and at complex interfaces.
- Development of microbial systems biology for bioenergy applications.
- Measurement of the effects of microbes and viruses on the soil carbon cycle.
- Creation of secure biosystem design tools for environmental biocontainment.
- Development of new bioimaging techniques.
LLNL BER Programs
Program for Climate Model Diagnosis and Intercomparison (PCMDI) Center for Earth System Model Analysis (CESMA)
PCMDI-CESMA develops advanced methods and tools for the intercomparison of models that simulate the global Earth system's variability and change over time scales from decades to centuries. Current projects focus on supporting the intercomparison of modeling results from every major international climate modeling center; standardized metrics of model scientific performance; identifying robust cloud feedbacks in observations and models; and devising robust statistical methods for Earth system predictability.
Energy Exascale Earth System Model (E3SM)
The E3SM project is an ongoing, multi-institution effort to develop a state-of-the-science Earth system modeling, simulation, and prediction model that optimizes the use of DOE laboratory resources. The E3SM model simulates the fully coupled Earth system at high resolution. It incorporates coupling among energy, water, land-use, and related energy-relevant activities with a focus on near-term hind-casts for model validation and a near-term projection for energy sector planning. A major motivation for the E3SM project is the paradigm shift in computing architectures and their related programming models as computational capabilities move towards the exascale era. E3SM is optimizing code performance for current and next-generation DOE computer facilities.
Cloud Feedbacks
To reduce uncertainties associated with climate change, a team of LLNL and University of California, Los Angeles, researchers are using observations to determine which of the climate-model predicted responses of clouds to climate change are realistic and predictable.
Learn more about cloud feedbacks
Biogeochemistry at Interfaces
LLNL’s Biogeochemistry at Interfaces program is developing conceptual and numerical models of metal and radionuclide cycling and mobility in the subsurface environment. Our focus is identifying biogeochemical processes at environmentally relevant timescales and at biogeochemically complex interfaces (i.e., gradients in salinity and redox chemistry in estuary and pond sediments) at field sites across the U.S. The program is identifying coupled processes—such as microbial community composition, iron redox chemistry and mineral dissolution/precipitation—that can stabilize metals and radionuclides in the subsurface at environmentally relevant timescales.
Collaborators: Clemson University, University of Florida, University of Helsinki, Karlsruhe Institute of Technology, Lawrence Berkeley National Laboratory, Pacific Northwest National Laboratory, and Savannah River National Laboratory.
Bioenergy
Photosynthetic algal and plant systems convert solar energy and carbon dioxide into useful organic molecules. Their growth and efficiency are shaped and assisted by the microbial communities that dwell in and around them and live off their primary productivity. The LLNL Bioenergy Scientific Focus Area focuses on the community systems biology of microbial consortia closely associated with bioenergy-relevant plants and algae, with the ultimate goal of developing predictive frameworks to understand how these systems will respond to external stimuli. The project is uncovering new mechanisms of phototroph-symbiont interactions and how these interactions shape biomass productivity, crop robustness, the balance of resource fluxes (carbon, nutrients, water), and the functionality of the surrounding microbiome.
Collaborators: Massachusetts Institute of Technology, University of California at Berkeley, North Carolina State University, Lawrence Berkeley National Laboratory, and Sandia National Laboratories.
Soil Microbiomes
LLNL’s Soil Microbiome Scientific Focus Area—Microbes Persist: Systems Biology of the Soil Microbiome—seeks to understand how microbial ecophysiology, population dynamics and microbe–mineral–organic matter interactions regulate the persistence of microbial residues in soil. The project has revealed functions and interactions of soil microorganisms and viruses that cannot be cultivated and has contributed to a new understanding of how environmental factors shape carbon stored in soil. We are applying our unique capabilities in isotope tracing combined with ‘omics’ approaches to study individual genomes, expression and metabolic products of active microbes within their natural habitat. These capabilities include LLNL’s High-Throughput Stable Isotope Probing pipeline, in-house informatics tools for virome and metagenome analysis, high-resolution NanoSIMS isotopic imaging, and carbon-14 accelerator mass spectrometry.
Collaborators: University of California at Berkeley, Northern Arizona University, Ohio State University, Lawrence Berkeley National Laboratory, and the Pacific Northwest National Laboratory.
Secure Biosystems Design
LLNL’s Biosecurity SFA focuses on building layered safeguard mechanisms at sequence, cellular and population levels. Plant benefiting Pseudomonads are engineered and tested for containment and function in relevant soil and rhizosphere environment, delivering effective safeguard mechanisms that can be applied to a broad range of microorganisms. The program couples machine learning (ML) with high-throughput assays to uncover key design parameters of sequence entanglement to increase gene stability and mitigate horizontal gene transfer. LLNL capabilities in ML-enabled protein design, high throughput protein structure modeling, multiplexed gene synthesis, pooled gene library screening, genetic circuit designs, and microfluidics with spatial and temporal controls enable an iterative secure biosystems design strategy of design, build, test, and learn.
Collaborators: Columbia University, University of Maryland, University of Minnesota, and University of California at Berkeley.
Biological Imaging Capabilities
Rhizosphere microscope
We are building a light microscope to enable live, long-term imaging of plant-microbe-mineral interactions in the rhizosphere to study carbon cycling, sustainable food, and fuel production and environmental processes, including contaminant transport. We are using adaptive optics (AO) and label-free multiphoton microscopy and spectroscopy to overcome the challenges of optical imaging in mineral and soil matrices. Multimodal detection will enhance label-free visualization of microbial cells and organic matter.
NanoSIMS Isotopic Imaging
Quantifying microbial function in complex communities is central to the Office of Science’s mission to develop a predictive understanding of microbial function for bioenergy, carbon sequestration, and environmental decontamination. LLNL initiated single-cell isotope tracing for DOE BER microbial ecology studies in 2003. We combine stable isotope probing (SIP) with isotopic imaging using the LLNL NanoSIMS 50—an approach that we call “nanoSIP” (Pett-Ridge and Weber, 2012). Typically, nanoSIP analyses are combined with geochemical, genomic, and other omics data. Fluorescence in situ hybridization (FISH) can be used to link identity to function.
3D Quantum Microscope
We are developing a new 3D imaging modality that uses quantum entangled photon pairs to obtain more information about fluorescence and scattering events than is available in standard fluorescence or scattering measurements. The entangled photons will enable our microscope to use two separate 2D detectors to obtain 3- and 4-dimensional information about the same photon absorption/fluorescence emission event or scattering event in the sample. Based on this concept, we envision a new, 3D Quantum (3DQ) microscope that uses quantum entangled light to provide 3D optical imaging at high frame rates. We will apply this microscope to dynamic host-bacterial interactions in bioenergy algal pond and plant systems.
Collaborators: École Polytechnique Fédérale de Lausanne and Colorado School of Mines.
Felice Lightstone
Lab Program Coordinator for BER at LLNL
felice [at] llnl.gov (felice[at]llnl[dot]gov)